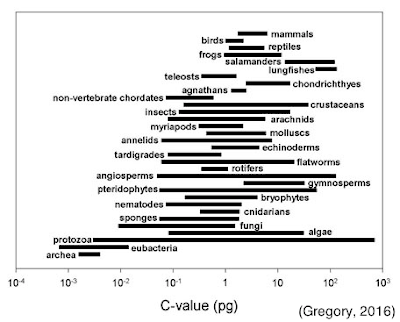
One aspect of biology that has often troubled scientists and students
alike is referred to, now somewhat colloquially, as the C-Value paradox. The C-Value specifically refers to total DNA
content, not chromosome number, but the two are often related. It seems intuitive to expect that the more
‘complex’ an organism, the more chromosomes or total DNA it should have. This has led to the erroneous assumption that
things like mammals, from our biased perspective, should have high DNA content. But it turns out that this idea is far from the
truth, by several orders of magnitude.
In fact, the organism with the highest chromosome number out of anything
living today is a fern, Ophioglossum
reticulatum1, with a diploid count of 2n=1440 (Ghatak, 1977). And this species isn’t
even a particularly complex fern either; it produces only one leaf, grows to a
maximum height of 15 cm (~5 in), and spends a significant portion of its
lifecycle underground. But ferns in general
tend to have high chromosome counts2, even when compared to other
plants, with an average haploid count of n=57.05 (Klekowski and Baker,
1966). So what do ferns have going for
them that we don’t, and what are all those extra chromosomes being used for?
These
types of questions were first asked back in the 1950s and -60s when it was
discovered that different organisms contained different amounts of DNA and that
some organisms considered to be ‘primitive,’ such as fish and amphibians,
actually contain 20X as much DNA as humans.
This fact didn’t sit well with everyone, especially those that viewed Homo sapiens as the pinnacle of
evolution. One author even lamented
that, “the lowly liverwort has 18 times as much DNA as we, and the slimy, dull
salamander known as Amphiuma has 26 times our complement
of DNA” (Comings, 1972).
C.A. Thomas, Jr.
dubbed this disparity between the genome sizes of different organisms the
C-value paradox, in which ‘C-value’ stands for the total amount of DNA in a
haploid set of chromosomes (Thomas, Jr., 1971).
Sometimes even closely related species can range widely in their DNA
content, such as an 80-fold difference in the buttercups (Ranunculaceae) and as
much as a 2000-fold difference in some algae (Rothfels et al., 1966;
Holm-Hansen, 1969). There are a number
of different mechanisms underlying this phenomenon, but we’ll focus on just
one: polyploidy is a phenomenon that affects plants more than any other
organism, and affects ferns more than any other group of plants.
Polyploidy is
simply the state of having more than the ‘normal’ two sets of chromosomes (one
from mom and one from dad). There can be
triploids (3 sets), tetraploids (4), hexaploids (6) all the way up to Ophioglossum reticulatum, which has about 96 sets of chromosomes (nonahexaploid)! If this sounds
amazing to you, it does to biologists as well.
Even the renowned Stephen Jay Gould (1991) admitted to being at a loss
for words when he wrote about this fern some twenty-five years ago, and G. L.
Stebbins (1971), one of the great botanists of the 20th century,
wrote that it is “nothing short of miraculous” the species could routinely and
accurately undergo meiosis with such high numbers of chromosomes.
One of the reasons
polyploidy is so commonplace in plants is it allows them to overcome the
problem of sterility after hybridization.
When two plants that are distantly enough related fertilize and create a
hybrid, the genetic material of the two parents is often too dissimilar to
successfully pair during meiosis. We see
this in the animal kingdom too, the most well-known example probably being that
of the sterile mule (horse X donkey).
Polyploidy allows for a quick fix to this problem in plants,
however. If the hybrid undergoes
chromosome doubling, then now each chromosome has an exact copy of itself to
pair with during meiosis, and fertility is automatically restored. In other words, life, as it were, finds a
way. But obviously mules haven’t been
able to take advantage of this solution, which is because polyploidy is generally
not observed as much in animals as it in plants, although this statement is not
without exception. It’s not that
chromosome doubling doesn’t occur, but instead that most animals don’t seem well
equipped to cope with the physiological consequences of having several copies
of each gene.
The study
of polyploidy encapsulates the entire tree of life and contains numerous facets
that scientists have explored, such as how genes on duplicated chromosomes can
be co-opted to code for new traits, or how species often revert back to a
diploid state after chromosome doubling.
And if you were to go in the opposite direction, to find the organism with
the least number of chromosomes, you’d need look no further than the ant Myrmecia pilosula, in which the males,
who develop from unfertilized eggs, have just one chromosome (Crosland and
Crozier, 1986).
1This is not
counting the protist Oxytricha trifallax,
which has “arguably the most complex genome architecture of any known
eukaryote” (Miller, 2014). This organism
has an astounding 16,000 nanochromosomes, which are tiny and often only
comprised of one gene (Swart et al., 2014).
2True for homosporous
ferns; heterosporous ferns, which are exclusively aquatic, actually have
comparatively low chromosome numbers, and no one really knows why.
Citations
Abraham, A., & Ninan, C. A. (1954).
Chromosomes of Ophioglossum reticulatum L. Current
Science 23: 213-214.
Comings, D. E.
(1972). The structure and function of chromatin. Advance human
genetics
3: 237-431.
Crosland, M. W., & Crozier, R. H.
(1986). Myrmecia pilosula, an ant with only one pair
of chromosomes. Science 231: 1278-1278.
Ghatak,
J. (1977). Biosystematic survey of the pteridophytes from Shevaroy Hills, South
India. Nucleus, Calcutta 20: 105-108.
natural
history. New York: Norton.
Gregory, T. R. (2016). Animal Genome
Size Database. http://www.genomesize.com.
Holm-Hansen, O.
(1969). Algae: amounts of DNA and organic carbon in single cells.
Science,
163: 87-88.
Klekowski, E. J.,
& Baker, H. G. (1966). Evolutionary significance of polyploidy in the
Pteridophyta. Science, 153: 305-307.
Masterson, J.
(1994). Stomatal size in fossil plants: evidence for polyploidy in
majority
of angiosperms. Science-AAAS-Weekly Paper Edition-including Guide to
Scientific Information, 264(5157), 421-423.
Miller, G. (2014, September). This Bizarre Organism Builds Itself a New Genome Every
Time It Has Sex. Retrieved January 21, 2016, from
http://www.wired.com/2014/09/oxytricha-encrypted-genome/
Pray, L. & Zhaurova, K. (2008) Barbara
McClintock and the discovery of jumping
genes (transposons). Nature Education 1:169
Rothfels, K.,
Sexsmith, E., Heimburger, M., & Krause, M. O. (1966). Chromosome size
and
DNA content of species of Anemone L. and related genera (Ranunculaceae). Chromosome, 20: 54-74.
Stebbins GL. 1971. Chromosome evolution
in higher plants. London: Edward Arnold.
Swart, E. C., Bracht, J. R., Magrini, V., Minx,
P., Chen, X., Zhou, Y., ... & Jung, S. (2013). The Oxytricha trifallax
macronuclear genome: a complex eukaryotic genome with 16,000 tiny
chromosomes. PLoS Biol, 11(1), e1001473.
Wood, T. E., Takebayashi, N., Barker, M. S., Mayrose, I., Greenspoon, P. B., & Rieseberg, L. H. (2009). The frequency of polyploid speciation in vascular plants. Proceedings of the national Academy of sciences 106: 13875-13879.
Wood, T. E., Takebayashi, N., Barker, M. S., Mayrose, I., Greenspoon, P. B., & Rieseberg, L. H. (2009). The frequency of polyploid speciation in vascular plants. Proceedings of the national Academy of sciences 106: 13875-13879.